Distribution Classes#
The Distribution classes define the methods for generating particle sizes used in the simulation. These classes provide flexibility in modeling particles of varying sizes according to different statistical distributions, such as normal, log-normal, Weibull, and more. This allows for realistic simulation of particle populations with varying size characteristics.
Available Distribution Classes#
The following are the available particle size distribution classes in FlowCyPy:
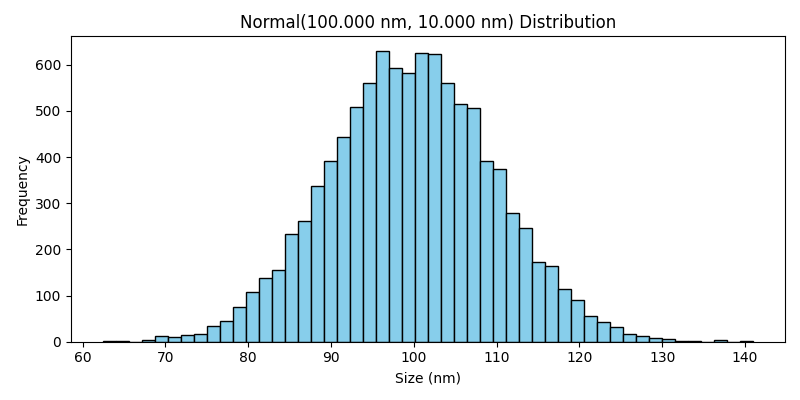
Normal Distribution#
The Normal class generates particle sizes that follow a normal (Gaussian) distribution. This distribution is characterized by a symmetric bell curve, where most particles are concentrated around the mean size, with fewer particles at the extremes.
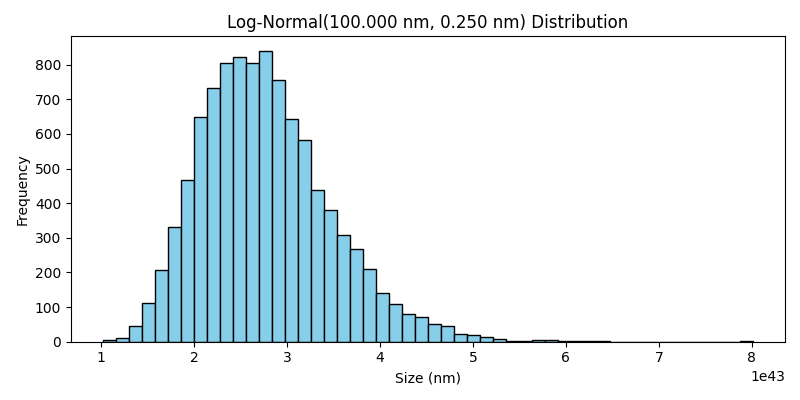
LogNormal Distribution#
The LogNormal class generates particle sizes based on a log-normal distribution. In this case, the logarithm of the particle sizes follows a normal distribution. This type of distribution is often used to model phenomena where the particle sizes are positively skewed, with a long tail towards larger sizes.
Rosin-Rammler Distribution#
The RosinRammler class generates particle sizes using the Rosin-Rammler distribution, which is commonly used to describe the size distribution of powders and granular materials. It provides a skewed distribution where most particles are within a specific range, but some larger particles may exist.
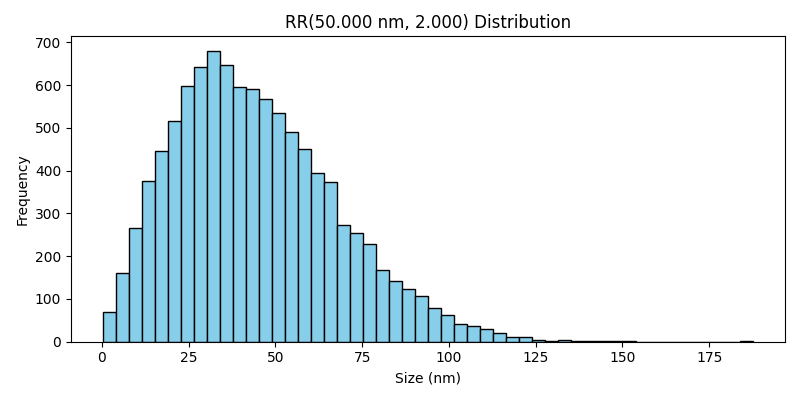
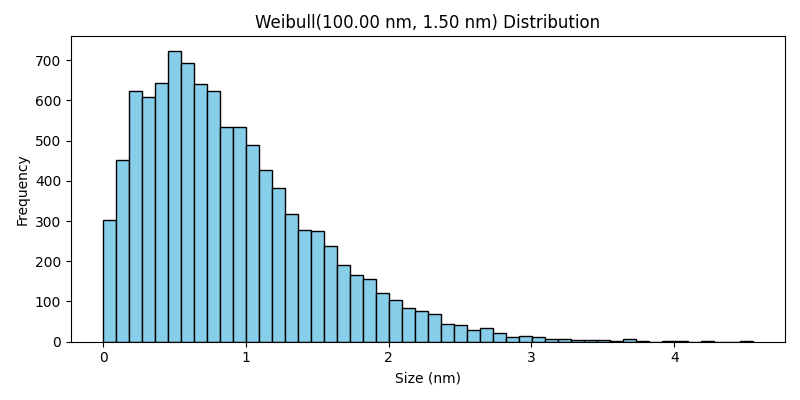
Weibull Distribution#
The Weibull class generates particle sizes according to the Weibull distribution. This distribution is flexible and can model various types of particle size distributions, ranging from light-tailed to heavy-tailed distributions, depending on the shape parameter.
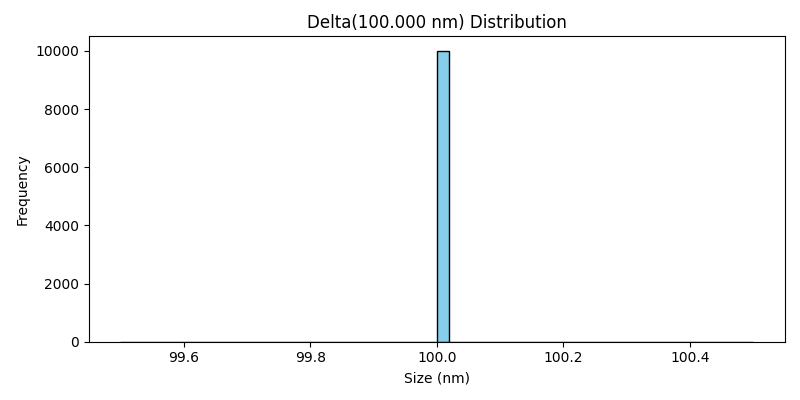
Delta Distribution#
The Delta class models particle sizes as a delta function, where all particles have exactly the same size. This distribution is useful for simulations where all particles are of a fixed size, without any variation.
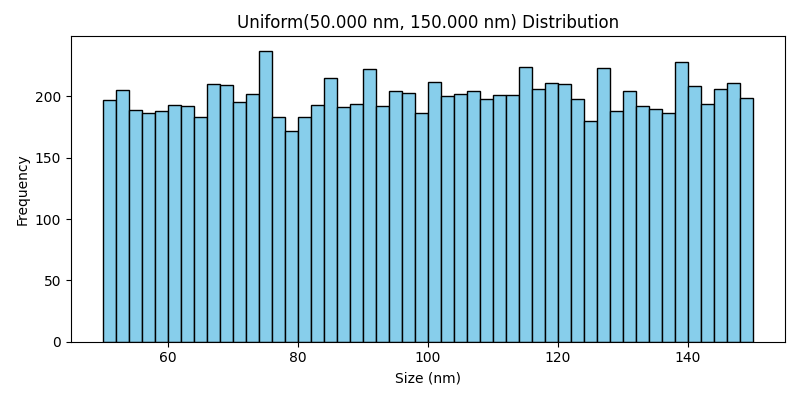
Uniform Distribution#
The Uniform class generates particle sizes that are evenly distributed between a specified lower and upper bound. This results in a flat distribution, where all particle sizes within the range are equally likely to occur.