Note
Go to the end to download the full example code.
WorkFlow#
This script simulates flow cytometer signals using the FlowCytometer class and analyzes the results using the PulseAnalyzer class from the FlowCyPy library. The signals generated (forward scatter and side scatter) provide insights into the physical properties of particles passing through the laser beam.
Workflow:#
Define a particle diameter distribution using ScattererCollection.
Simulate flow cytometer signals using FlowCytometer.
Analyze the forward scatter signal with PulseAnalyzer to extract features like peak height, width, and area.
Visualize the generated signals and display the extracted pulse features.
Step 1: Import necessary modules from FlowCyPy
from FlowCyPy import FlowCytometer, ScattererCollection, Detector, GaussianBeam, TransimpedanceAmplifier
from FlowCyPy.flow_cell import FlowCell
from FlowCyPy import distribution
from FlowCyPy.population import Sphere
from FlowCyPy.signal_digitizer import SignalDigitizer
from FlowCyPy import units
Step 2: Define the laser source#
Set up a laser source with a wavelength of 1550 nm, optical power of 200 mW, and a numerical aperture of 0.3.
source = GaussianBeam(
numerical_aperture=0.3 * units.AU, # Numerical aperture: 0.3
wavelength=800 * units.nanometer, # Laser wavelength: 800 nm
optical_power=20 * units.milliwatt # Optical power: 20 milliwatts
)
Step 3: Define flow parameters#
Set the flow speed to 80 micrometers per second and a flow area of 1 square micrometer, with a total simulation time of 1 second.
flow_cell = FlowCell(
sample_volume_flow=0.02 * units.microliter / units.second, # Flow speed: 10 microliter per second
sheath_volume_flow=0.1 * units.microliter / units.second, # Flow speed: 10 microliter per second
width=20 * units.micrometer, # Flow area: 10 x 10 micrometers
height=10 * units.micrometer, # Flow area: 10 x 10 micrometers
)
Step 4: Define the particle diameter distribution#
Use a normal diameter distribution with a mean diameter of 200 nanometers and a standard deviation of 10 nanometers. This represents the population of scatterers (particles) that will interact with the laser source.
ev_diameter = distribution.Normal(
mean=200 * units.nanometer, # Mean particle diameter: 200 nanometers
std_dev=10 * units.nanometer # Standard deviation: 10 nanometers
)
ev_ri = distribution.Normal(
mean=1.39 * units.RIU, # Mean refractive index: 1.39
std_dev=0.01 * units.RIU # Standard deviation: 0.01
)
ev = Sphere(
particle_count=10 * units.particle,
diameter=ev_diameter, # Particle diameter distribution
refractive_index=ev_ri, # Refractive index distribution
name='EV' # Name of the particle population: Extracellular Vesicles (EV)
)
scatterer_collection = ScattererCollection()
scatterer_collection.add_population(ev)
# Step 5: Define the detector
# ---------------------------
# The detector captures the scattered light. It is positioned at 90 degrees relative to the incident light beam
# and configured with a numerical aperture of 0.4 and responsivity of 1.
digitizer = SignalDigitizer(
bit_depth=1024,
saturation_levels='auto',
sampling_rate=1 * units.megahertz, # Sampling frequency: 1 MHz
)
detector_0 = Detector(
phi_angle=90 * units.degree, # Detector angle: 90 degrees (Side Scatter)
numerical_aperture=0.4 * units.AU, # Numerical aperture of the detector
name='first detector', # Detector name
responsivity=1 * units.ampere / units.watt, # Responsitivity of the detector (light to signal conversion efficiency)
)
detector_1 = Detector(
phi_angle=0 * units.degree, # Detector angle: 90 degrees (Sid e Scatter)
numerical_aperture=0.4 * units.AU, # Numerical aperture of the detector
name='second detector', # Detector name
responsivity=1 * units.ampere / units.watt, # Responsitivity of the detector (light to signal conversion efficiency)
)
transimpedance_amplifier = TransimpedanceAmplifier(
gain=100 * units.volt / units.ampere,
bandwidth = 10 * units.megahertz
)
# Step 6: Simulate Flow Cytometer Signals
# ---------------------------------------
# Create a FlowCytometer instance to simulate the signals generated as particles pass through the laser beam.
cytometer = FlowCytometer(
source=source,
transimpedance_amplifier=transimpedance_amplifier,
digitizer=digitizer,
scatterer_collection=scatterer_collection,
flow_cell=flow_cell, # Particle diameter distribution
detectors=[detector_0, detector_1] # List of detectors used in the simulation
)
# Run the flow cytometry simulation
cytometer.prepare_acquisition(run_time=0.2 * units.millisecond)
acquisition = cytometer.get_acquisition()
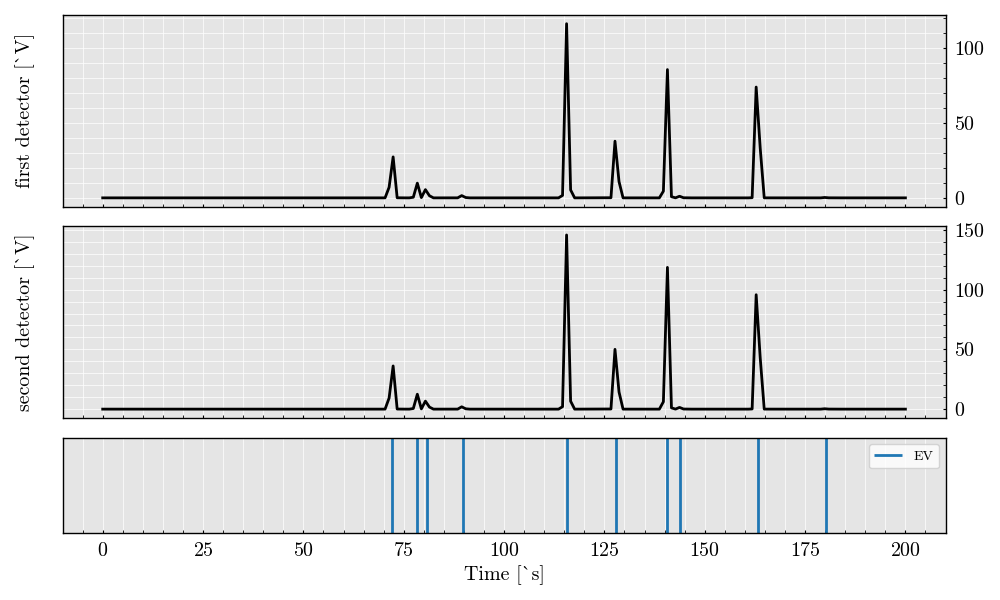
# Visualize the scatter signals from both detectors
acquisition.plot()
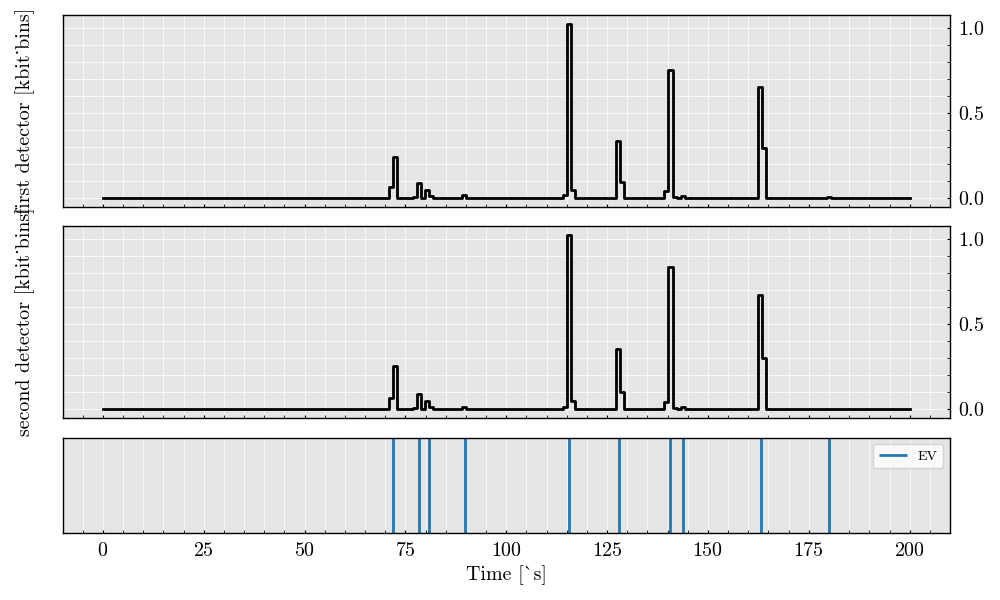
digital_signals = acquisition.digitalize(digitizer=digitizer)
digital_signals.plot()
"""
Summary:
--------
This script simulates flow cytometer signals, processes them to detect peaks in the forward scatter channel,
and extracts important features. The process is visualized through signal plots, and key properties are displayed.
"""
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/FlowCyPy/source.py:269: UserWarning: Transverse distribution of particle flow exceed the waist of the source
warnings.warn('Transverse distribution of particle flow exceed the waist of the source')
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/FlowCyPy/source.py:269: UserWarning: Transverse distribution of particle flow exceed the waist of the source
warnings.warn('Transverse distribution of particle flow exceed the waist of the source')
'\nSummary:\n--------\nThis script simulates flow cytometer signals, processes them to detect peaks in the forward scatter channel,\nand extracts important features. The process is visualized through signal plots, and key properties are displayed.\n'
Total running time of the script: (0 minutes 0.945 seconds)