Note
Go to the end to download the full example code.
Limit of Detection#
This example simulates the detection of small nanoparticles (90–150 nm diameter) in a flow cytometry setup using a dual-detector configuration (side and forward scatter). The simulation includes noise models, realistic fluidics, analog signal conditioning, digitization, triggering, and peak detection.
The main goal is to evaluate whether such particles produce detectable and distinguishable scatter signals in the presence of system noise and fluidic variability.
import numpy as np
from TypedUnit import ureg
from FlowCyPy import FlowCytometer, SimulationSettings
from FlowCyPy.fluidics import (
FlowCell,
Fluidics,
ScattererCollection,
distribution,
population,
)
from FlowCyPy.opto_electronics import (
Detector,
OptoElectronics,
TransimpedanceAmplifier,
source,
)
from FlowCyPy.signal_processing import (
Digitizer,
SignalProcessing,
circuits,
peak_locator,
triggering_system,
)
Simulation Configuration
SimulationSettings.include_noises = True
SimulationSettings.include_shot_noise = True
SimulationSettings.include_source_noise = True
SimulationSettings.include_dark_current_noise = True
SimulationSettings.assume_perfect_hydrodynamic_focusing = True
np.random.seed(3)
Optical Source
source = source.GaussianBeam(
numerical_aperture=0.2 * ureg.AU,
wavelength=405 * ureg.nanometer,
optical_power=100 * ureg.milliwatt,
)
Flow Cell Configuration
flow_cell = FlowCell(
sample_volume_flow=0.02 * ureg.microliter / ureg.second,
sheath_volume_flow=0.1 * ureg.microliter / ureg.second,
width=20 * ureg.micrometer,
height=10 * ureg.micrometer,
)
Define Scatterer Populations (90–150 nm spheres)
scatterer_collection = ScattererCollection()
for size in [150, 125, 100, 75, 50]:
pop = population.Sphere(
name=f"{size} nm",
particle_count=15 * ureg.particle,
diameter=distribution.Delta(position=size * ureg.nanometer),
refractive_index=distribution.Delta(position=1.36 * ureg.RIU),
medium_refractive_index=1.33 * ureg.RIU,
)
scatterer_collection.add_population(pop)
Fluidics Subsystem
fluidics = Fluidics(scatterer_collection=scatterer_collection, flow_cell=flow_cell)
Signal Digitizer
digitizer = Digitizer(
bit_depth="14bit", saturation_levels="auto", sampling_rate=60 * ureg.megahertz
)
Detectors
detector_side = Detector(
name="side",
phi_angle=90 * ureg.degree,
numerical_aperture=0.2 * ureg.AU,
responsivity=1 * ureg.ampere / ureg.watt,
dark_current=0.01 * ureg.milliampere,
)
detector_forward = Detector(
name="forward",
phi_angle=0 * ureg.degree,
numerical_aperture=0.2 * ureg.AU,
responsivity=1 * ureg.ampere / ureg.watt,
dark_current=0.01 * ureg.milliampere,
)
Amplifier and Opto-Electronics
amplifier = TransimpedanceAmplifier(
gain=10_000 * ureg.volt / ureg.ampere, bandwidth=10 * ureg.megahertz
)
opto_electronics = OptoElectronics(
detectors=[detector_side, detector_forward], source=source, amplifier=amplifier
)
Analog Processing Pipeline
analog_processing = [
circuits.BaselineRestorator(window_size=10 * ureg.microsecond),
circuits.BesselLowPass(cutoff=1.5 * ureg.megahertz, order=4, gain=2),
]
Triggering and Peak Detection
triggering_system = triggering_system.DynamicWindow(
trigger_detector_name="forward",
threshold="5sigma",
max_triggers=-1,
pre_buffer=128,
post_buffer=128,
)
signal_processing = SignalProcessing(
digitizer=digitizer,
analog_processing=analog_processing,
triggering_system=triggering_system,
peak_algorithm=peak_locator.GlobalPeakLocator(),
)
cytometer = FlowCytometer(
opto_electronics=opto_electronics,
fluidics=fluidics,
signal_processing=signal_processing,
background_power=0.0001 * ureg.milliwatt,
)
run_record = cytometer.run(run_time=5.0 * ureg.millisecond)
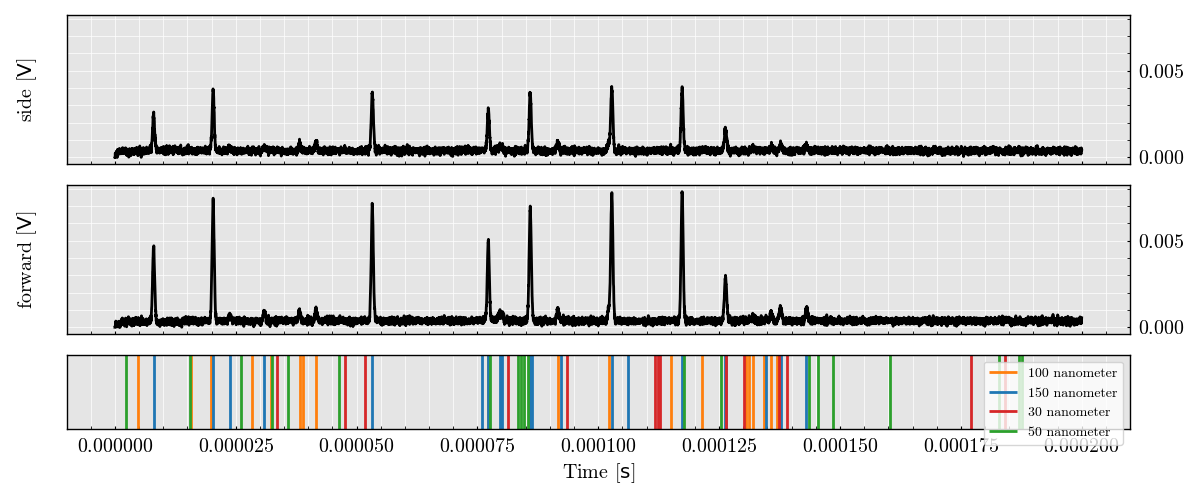
Plot Raw Analog Signal
run_record.plot_analog()
<Figure size 800x500 with 3 Axes>
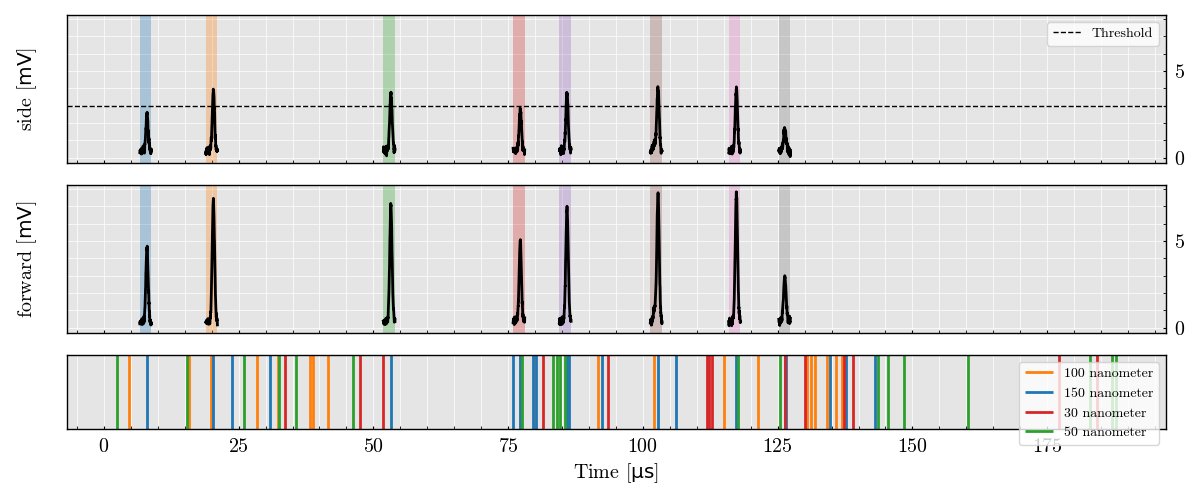
Plot Triggered Analog Signal Segments
run_record.plot_digital()
<Figure size 800x500 with 3 Axes>
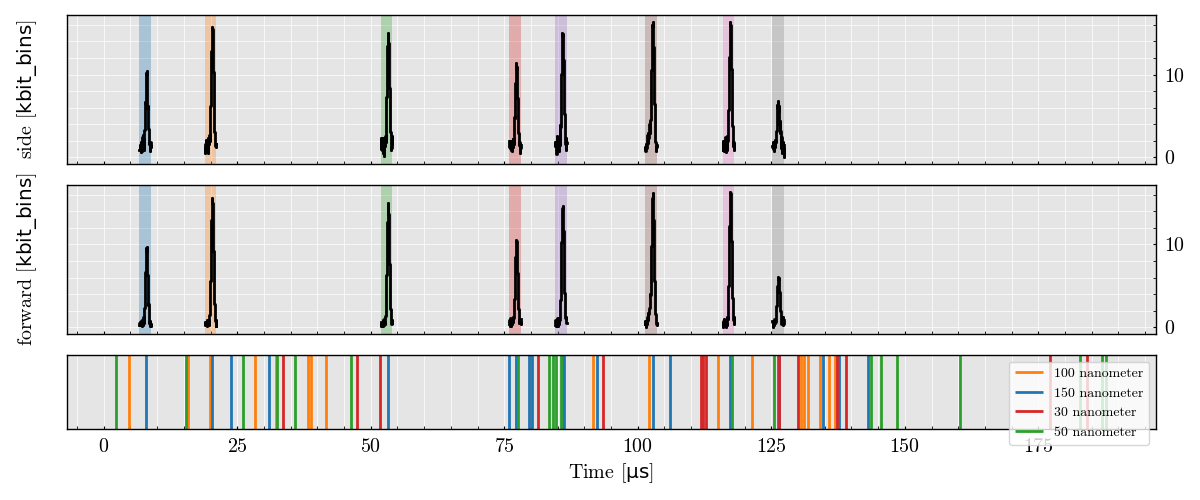
Plot Peak Features (Side vs Forward Height)
run_record.peaks.plot(x=("side", "Height"), y=("forward", "Height"))
<Figure size 600x600 with 3 Axes>
Total running time of the script: (0 minutes 13.124 seconds)