Note
Go to the end to download the full example code.
Workflow#
This tutorial demonstrates how to simulate a flow cytometry experiment using the FlowCyPy library. The simulation involves configuring a flow setup, defining a single population of particles, and analyzing scattering signals from two detectors to produce a 2D density plot of scattering intensities.
Overview:#
Configure the flow cell and particle population.
Define the laser source and detector parameters.
Simulate the flow cytometry experiment.
Analyze the generated signals and visualize results.
Step 0: Import Necessary Libraries#
Here, we import the necessary libraries and units for the simulation. The units module helps us define physical quantities like meters, seconds, and watts in a concise and consistent manner.
from FlowCyPy import units
from FlowCyPy import GaussianBeam
from FlowCyPy.flow_cell import FlowCell
from FlowCyPy import ScattererCollection
from FlowCyPy.population import Exosome, HDL
from FlowCyPy.detector import PMT
from FlowCyPy.signal_digitizer import SignalDigitizer
from FlowCyPy import FlowCytometer, TransimpedanceAmplifier
source = GaussianBeam(
numerical_aperture=0.3 * units.AU, # Numerical aperture
wavelength=200 * units.nanometer, # Wavelength
optical_power=20 * units.milliwatt # Optical power
)
flow_cell = FlowCell(
sample_volume_flow=0.02 * units.microliter / units.second,
sheath_volume_flow=0.1 * units.microliter / units.second,
width=20 * units.micrometer,
height=10 * units.micrometer,
)
scatterer_collection = ScattererCollection(medium_refractive_index=1.33 * units.RIU)
# Add an Exosome and HDL population
scatterer_collection.add_population(
Exosome(particle_count=5e9 * units.particle / units.milliliter),
HDL(particle_count=5e9 * units.particle / units.milliliter)
)
scatterer_collection.dilute(factor=1)
# Initialize the scatterer with the flow cell
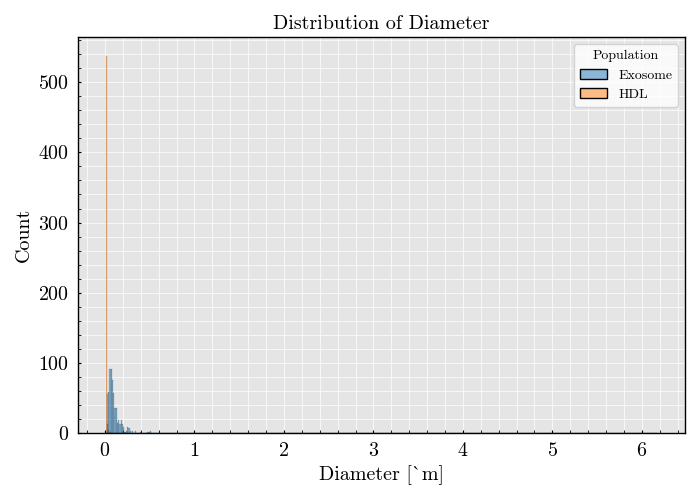
df = scatterer_collection.get_population_dataframe(total_sampling=600, use_ratio=False) # Visualize the particle population
df.plot(x='Diameter', bins='auto')
digitizer = SignalDigitizer(
bit_depth='14bit',
saturation_levels='auto',
sampling_rate=60 * units.megahertz,
)
detector_0 = PMT(name='forward', phi_angle=0 * units.degree, numerical_aperture=0.3 * units.AU)
detector_1 = PMT(name='side', phi_angle=90 * units.degree, numerical_aperture=0.3 * units.AU)
transimpedance_amplifier = TransimpedanceAmplifier(
gain=100 * units.volt / units.ampere,
bandwidth = 10 * units.megahertz
)
cytometer = FlowCytometer(
source=source,
transimpedance_amplifier=transimpedance_amplifier,
scatterer_collection=scatterer_collection,
digitizer=digitizer,
detectors=[detector_0, detector_1],
flow_cell=flow_cell,
background_power=0.001 * units.milliwatt
)
# Run the flow cytometry simulation
cytometer.prepare_acquisition(run_time=0.1 * units.millisecond)
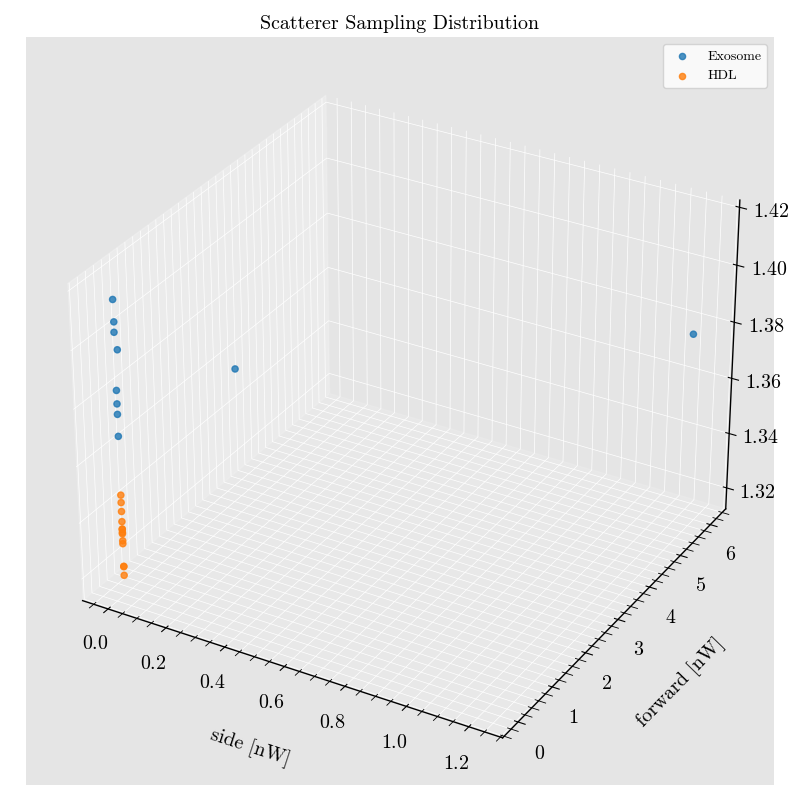
cytometer.scatterer_dataframe.plot(
x='side',
y='forward',
z='RefractiveIndex'
)
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/FlowCyPy/source.py:269: UserWarning: Transverse distribution of particle flow exceed the waist of the source
warnings.warn('Transverse distribution of particle flow exceed the waist of the source')
/opt/hostedtoolcache/Python/3.11.12/x64/lib/python3.11/site-packages/FlowCyPy/source.py:269: UserWarning: Transverse distribution of particle flow exceed the waist of the source
warnings.warn('Transverse distribution of particle flow exceed the waist of the source')
<Figure size 800x800 with 1 Axes>
Total running time of the script: (0 minutes 1.493 seconds)